Advanced metagenomics and RNA sequencing for diverse biomes: microbes, plants, animals, and aquatics
Biome- Genomics




Bacterial 16S (v1-v9) Amplicon metaSeq
To gain a deeper understanding of microbial communities and biodiversity present in various ecosystems
Fungal ITS Amplicon metaSeq
accurately identify and characterize the genetic profiles of microorganisms, shedding light on their ecological roles
Algal 18SrRNA Amplicon metaSeq
To uncover hidden diversity, track changes in algal communities over time, & assess the impacts of climate change & pollution

Shot-gun Whole genome -metaSeq
To unveil the complex interactions and metabolic capabilities of these microorganisms, enhancing our understanding of their roles in ecosystems, human health, & biotechnology
To explore un-characterized microbes, understanding their contributions to bio-geochemical cycles, and discovering novel enzymes and bio-active compounds
Metagenomic Assembled Genome- metaSeq
To explore the genetic diversity of un-culturable microorganisms in complex environmental samples
MAGs are reconstructed from mixed microbial communities, allowing for the identification of functional genes, metabolic pathways, & interactions among species
Meta-Transcriptomics - metaRNASeq
provides insights into the active metabolic processes occurring within microbial communities, helping us understand how these microorganisms adapt to and thrive in their environments





Next GEN RNA sequencing for diverse RNA's
RNAOmics
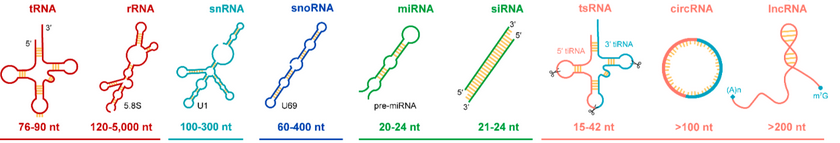
Provides in-depth insights into gene expression, transcript diversity & gene regulation
mRNAseq | Splice Varients
Encompasses the exploration of gene expression and transcript variability, providing insights into the complex mechanisms that drive cellular functions
Messenger RNA sequencing (mRNA seq)- To analyze the complete set of RNA transcripts produced by the genome at a specific moment in time, ultimately advancing our knowledge of gene regulation and function across various biological contexts
Provides in-depth insights in ncRNA landscape - ncRNAseq
Non-coding RNA (ncRNA) sequencing -
To understand their critical cellular activities like genomic imprinting, chromatin remodeling, & post-transcriptional regulation. Advances in sequencing technologies have allowed researchers
To explore the ncRNA landscape, unveiling their complexity and functional diversity
NGS solutions of Non-coding RNA landscape
- Explores the intricacies of ncRNAs in cellular process

Sample requirement
DNA amount ≥ 5 µg, concentration ≥ 100 ng/µL, 1.8 < OD260/280 < 2.0,2.0 < OD260/230 < 2.2
RNA amount ≥ 1 µg, concentration ≥ 50 ng/µL, RIN ≥ 8.0, OD260/280= ~2.0. OD260/230= 2.0 – 2.2
No detergents or surfactants in the buffer

Syngenome's innovative solutions transformed our approach to sustainable biomes and advanced our genomic research significantly.

The genomic insights provided by Syngenome have been invaluable for our synthetic biology projects and sustainability goals.

★★★★★
★★★★★
Innovation
Empowering sustainable biomes through advanced genomics solutions
© 2025 SYNGENOME (OPC) PRIVATE LIMITED | All rights reserved
Admin Unit:
Syngenome (OPC) Private Limited,
11-B4, Rajendra Nagar Extension, Narasimhanaickenpalayam, Coimbatore- 641 031, TN, India
info@syngenome.in +91-8667760889; +91-9486402305
Service - R & D Unit:
Syngenome (OPC) Private Limited,
171/A3, Second Floor, East Bashyakaralu Road, R.S.Puram, Coimbatore- 641 002, TN, India
infosyngenome@gmail.com +91-4223508993
Website Designed By: Syngenome


